
DR LEA LAHNSTEIN
Cross-Cutting Co-ordinator, Genomics England Clinical Interpretation Partnership
- The UK has sequenced 100,000 whole genomes in the National Health Service
- Collectively revolutionising medical practice and research, whole genome sequences are a resource that keeps on giving. Initially collected for patients and their clinical care, their digital nature makes them an ongoing resource for both research and the patients
- They are an example of the intricate relationship between technology and people
- Genomics is more fundamentally routed in the physical than other, more natively digital, realms of Big Data. This creates a unique set of requirements and infrastructure for Genomics
The ability of healthcare systems to deliver high-quality and ef cient care will increasingly depend on our ability to harness a growing wealth of data. We are all different from one another and the increasing accessibility of DNA sequencing is helping us characterise these differences. As in other elds, we are also increasingly connected to information, analytics and data collection, all powered by astounding leaps in computing power and data storage. Processes that used to take weeks now take hours on a single machine, which applies equally to DNA sequencing and deep machine learning. The general advent of the digital in healthcare is broad ranging, spanning machine learning algorithms for clinical decision-making, mobile apps, portals and patient monitoring.
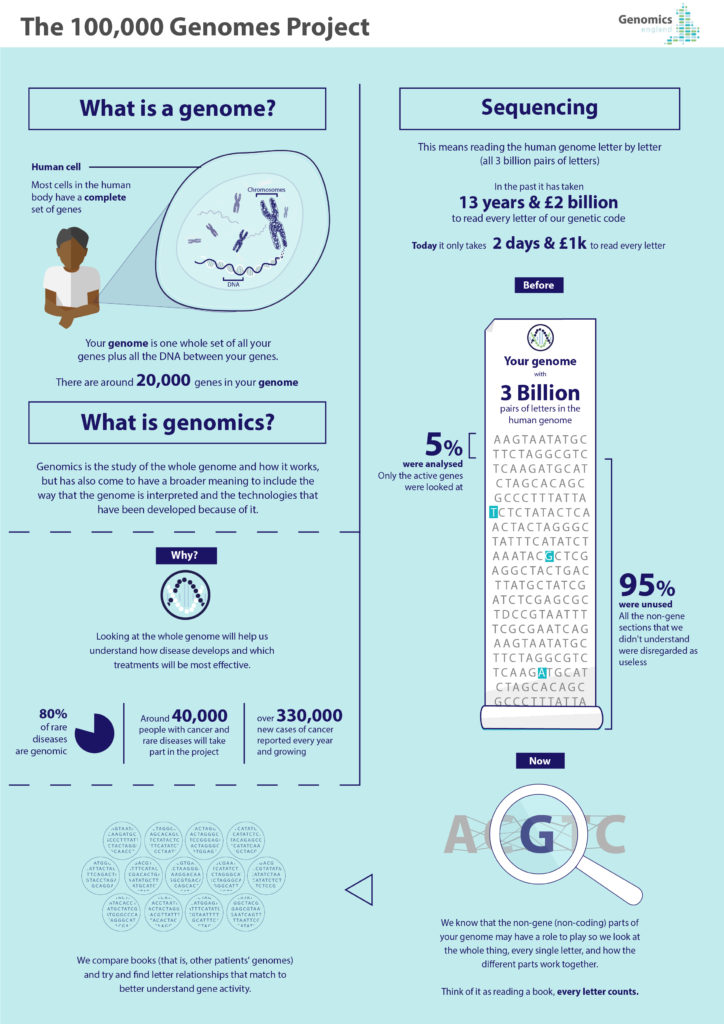
Genomic medicine uses individuals’ genomic information as part of their clinical care. Unlike specific genetic tests, the field of genomics focuses on the whole genome and how it works, including how it is to be interpreted and the technologies that have been developed to help do this. Genomics underpins the development of precision diagnostics, treatment and prevention strategies, helping to reduce side effects of drugs; target specific molecular changes through treatment; or predict how a disease will develop or how a drug might work.
For rare diseases, this means that, where clinicians might have difficulty telling symptoms apart and interpreting them based on observable characteristics alone, genomic testing can show up commonalities between seemingly different clinical pictures, or correct misdiagnoses based on superficially similar ones. For cancer, genomic and other biological research shows that no two tumours are the same. Cancer begins because of mutations in a normal cell’s DNA, developing mutations or changes that make it develop further; yet traditional cancer treatment is based mainly on statistical efficacy without being sure that there is a biological target in an individual patient’s cancer to act on.
New digital frontiers in genomics are increasingly allowing researchers and clinicians to spot the proverbial needle in the haystack of biological heterogeneity and compare what can work and what has worked in similar cases. They are also a learning curve for researchers, clinicians and patients alike and will shape – and be shaped by – their behaviours. In the UK, the 100,000 Genomes Project is employing digital technologies to harness the potential of genomic medicine1. Recently completed, this largest national sequencing project of its kind has (as of December 2018) sequenced an unprecedented 100,000 whole genomes2 from around 85,000 participants, who are National Health Service (NHS) patients with rare diseases, plus their families, and cancer patients.
Clinically, the effort is to find new diagnoses, treatment or disease management options for long-suffering rare disease3 patients and to find personalised treatment or clinical trial options for cancer4 patients. As well as being kept on file, genome sequences are analysed to compare patients’ genes against ones that are known to be associated with certain conditions and this analysis is beginning to include other variants in further parts of the genomes. The project has also transformed medicine and healthcare services forever and ushered in practices and lessons along the way. A new, consent-based, NHS Genomic Medicine Service is being created, transforming the way that patients are being cared for by offering new diagnoses and paving the way for more personalised, effective treatment options.
This is supported by further analysis of the genomes and their clinical interpretation by scientists, to generate new results and insights. Approved researchers can access5 the de-identified data generated by the project in a secure virtual and globally accessible Research Environment, where the combined genomes and rich clinical and health data held by the NHS for 85,000 people is a ground-breaking resource for new medical research. Commercial researchers are also able to join in the effort, kickstarting a UK genomics industry along the way.
This makes whole genome sequencing fundamentally different from other diagnostic tests, because we can keep looking for new answers in a participant’s genome, based on results coming in from other participants and based on new technology and algorithms becoming available. It is this process of opening up sequences for analysis and to researchers in order to “keep looking” that is almost entirely digital and based on the eld of Bioinformatics, the interdisciplinary field concerned with collecting and analysing large sets of complex biological data, such as genomic data, through specialised software tools and other methods. In order to enable this, the Research Environment, too, provides access in a digital space via secure log in.
The development and application of bioinformatic and genomic tools and technologies has long since become a field in its own right. It ranges from correctly assembling sequencing and comparing against reference data to find and evaluate tiny changes in them, to determining statistical evidence levels for new genomic tests. Crucially, however, the success of harnessing “the digital” to address genomic medicine for new diagnoses and insight is underpinned and even constituted by its relationship with everyone who comes into contact with it. In other clinical spaces, such as radiology, we see the importance of behavioural and social questions in the debates around machine learning, artificial intelligence and clinical decision-making. In this case, the clinical, technical, social and economic interpretation and utilisation of 100,000 whole genome sequences is unavoidably synonymous with certain principles and behaviours (this, too, is supported by designated research).
For those accessing the data (and for those governing that access), genomic research has to “think big”, speaking figuratively as well as literally. Research on the data from the 100,000 Genomes Project is organised through the Genomics England Clinical Interpretation Partnership (GeCIP)7, strictly controlling what can come in or out. A key part of the consent obtained from participants who donate their data is that no participant- level genomic or clinical data, or any other data that might identify them, can be removed from the environment. This means that all relevant resources and tools must be provided within the Research Environment and money, time and effort must be continually invested to keep the space fit for purpose and efficiently shared. Finally, genomic research, and the GeCIP in particular, allows for bolder research funding applications and landscape publications, again based on jointly staking out and tackling the unprecedented amount of data analysis opportunities afforded across the types of data and angles for analysing it. As with publicly funded storage and computing power, research dollars and publication inches are also likely to be allocated to collaborative and thus impactful efforts.
More fundamentally, genomic technologies are intertwined with the interests and behaviours of patients who stand to benefit from their application but who also allow for their data to be accessed for analysis and research. They underpin everything through their informed consent8 to take part in genomic medicine and donate their data to genomic research. Whole genome sequences differ from other diagnostic tests because the results they generate may emerge over a long time, as new technologies and further genomes analysed yield new insights, and some findings might apply to other conditions than the one that sequencing was commissioned for. This is particularly the case for the 100,000 Genomes Project, which is ground breaking in that it is a hybrid clinical and research project. Furthermore, since patterns and clues are continuously looked for across genomes to classify individual variation, genomic data are more likely to be useful to individuals the more information is available to be drawn on from others. Genomic technology binds us all together in contract, as the significance of a genetic variant for a particular condition or disease can only be evaluated in comparison with other genomes.
Ultimately, as impressive as genomic technology is, this space isn’t all digital but also very physical and visceral. To avoid the old “garbage in, garbage out” adage, this effort relies on setting up and following correct pathways for obtaining and handling tissue samples and clinical data. Most importantly, this is about real NHS patients with serious illnesses and about people living with conditions that impact their lives and influence their identity. They have been at the heart9 of the undertaking, supported it and ask for answers to their questions. They have to be physically present and they need physical access to genomic services.
This makes it so exciting that the 100,000 Genomes Project is being expanded so that at least one million genomes10 will now be sequenced and patients will continue to be given the option of donating their data to the research dataset. The future is now as the NHS Genomic Medicine Service11 will continue to embed genome sequencing in routine medical care and revolutionise healthcare by making it more digital, efficient and personalised.
Lea currently oversees the cross-cutting research strands for the Genomics England Clinical Interpretation Partnership (GeCIP) around the dataset of the 100,000 Genomes Project at Genomics England. She is responsible for constructing the framework for research in these areas, for conceiving and enacting the necessary governance and for ensuring that the necessary resources are available for researchers to fulfill their ambitions. Lea holds a longstanding interest in the intersection between biotechnology, bioscience and society, as well as in translating knowledge and expertise across multidisciplinary boundaries. After a PhD on the practices of biobanking and the exchange of biological samples and data, she was responsible at a cancer research and diagnostics company for projects encouraging the uptake of precision diagnostics through stakeholder management and information strategy, before working in Technology and Medical Innovation at GE Healthcare prior to her current role.
- https://www.genomicsengland.co.uk/ about-genomics-england/the-100000-ge- nomes-project/
- https://www.genomicsengland.co.uk/the- journey-to-100000-genomes/
- https://www.genomicsengland.co.uk/under- standing-genomics/jessicas-story/
- https://www.genomicsengland.co.uk/under- standing-genomics/8335-2/
- https://dtr.thalesesecurity.com/
- https://www.genomicsengland.co.uk/ about-gecip/for-gecip-members/data-and-da-ta-access/ [/_efn_note] for team science. GeCIP promotes collaboration in order to: avoid duplication of efforts and therefore computational and data storage requirements; bring together researchers across clinical and functional disciplines; cover the vast amount of ground required to analyse the huge amount of data needing to be integrated, harmonised for analysis and interpreted (1.6 billion clinical data points, millions of variants per genome and 21 Petabytes of data); and to create feedback loops between researchers and clinicians.
Participants’ privacy and anonymity is protected by sealing off the Research Environment6https://www.genomicsengland.co.uk/ about-genomics-england/research-environ- ment/
- https://www.genomicsengland.co.uk/ about-genomics-england/the-100000-ge- nomes-project/information-for-gmc-staff/ consent/
- https://www.genomicsengland.co.uk/ about-genomics-england/how-we-work/pa- tient-and-public-involvement/
- https://www.genomicsengland.co.uk/matt- hancock-announces-5-million-genomes-with- in- ve-years/
- https://www.england.nhs.uk/genomics/ nhs-genomic-med-service/



